NORLUX Neuro-Oncology Laboratory
NORLUX is an interdisciplinary preclinical research group dedicated to exploring brain tumor biology towards identification of innovative biomarkers and novel treatment options for patients.
activities
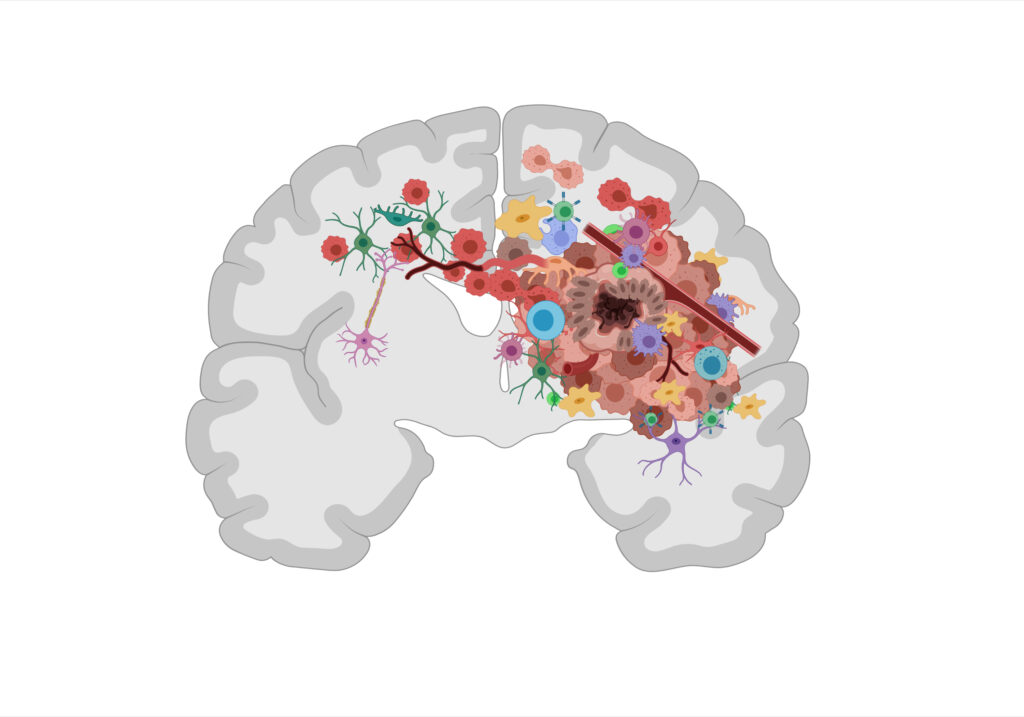
We are dedicated to advancing treatment options for malignant brain tumors, with a specific focus on diffuse gliomas. With a radically translational approach to our research, we employ patient samples and innovative patient-derived preclinical model systems that faithfully reproduce the patient tumor. In our research, we investigate the cellular and molecular basis of gliomas, we explore their capacity to invade healthy brain tissue and escape therapy. Our overarching goal is to reveal novel therapeutic targets and identify reliable biomarkers of treatment responses.
SCIENTIFIC AREAS OF INTEREST AND ACTIVITIES
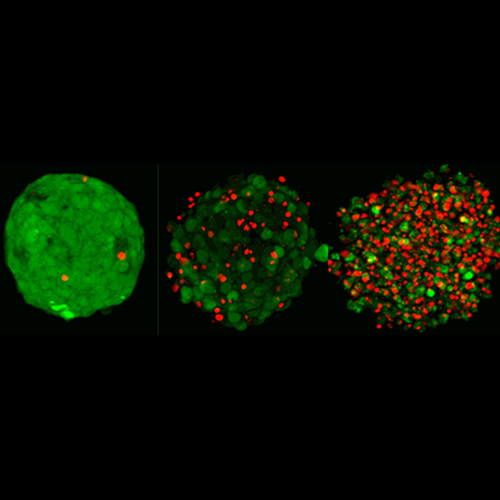
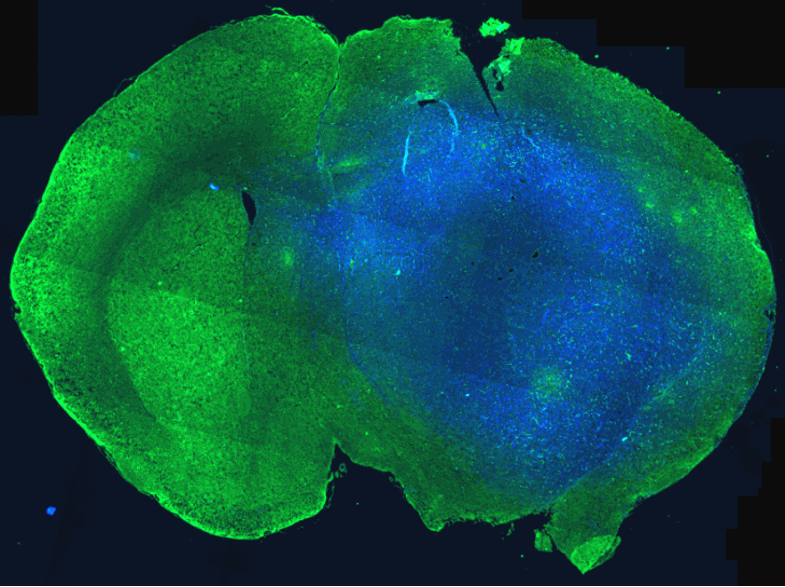
- Advanced brain tumor patient avatars for preclinical investigations and treatment efficacy studies
- Personalized medicine and pharmacogenomics
- Tumor heterogeneity at cellular, multi-omics and functional levels
- Tumor plasticity and resistance mechanisms
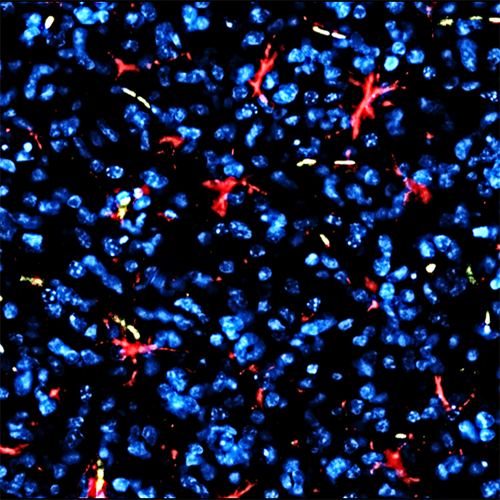
- Tumor-microenvironment interactions and immuno-oncology

Golebiewska
Patient cohorts and consortia
Funding Support
Projects & clinical trials
Featured team members
Scientific publications
-
Glioblastoma-instructed microglia transition to heterogeneous phenotypic states with phagocytic and dendritic cell-like features in patient tumors and patient-derived orthotopic xenografts – 02/04/2024
-
Corrigendum to ‘Molecular Identity Changes of Tumor-Associated Macrophages and Microglia After Magnetic Resonance Imaging–Guided Focused Ultrasound–Induced Blood–Brain Barrier Opening in a Mouse Glioblastoma Model’ [Ultrasound in Med & Biol. 49 (2023) 1082-1090, (S0301562922006664), (10.1016/j.ultrasmedbio.2022.12.006)] – 01/01/2024
-
PARK7/DJ-1 deficiency impairs microglial activation in response to LPS-induced inflammation – 09/02/2024
-
AllergoOncology – 01/01/2024
-
Magnetic resonance imaging-guided intracranial resection of glioblastoma tumors in patient-derived orthotopic xenografts leads to clinically relevant tumor recurrence – 02/01/2024
-
The epigenetic evolution of glioma is determined by the IDH1 mutation status and treatment regimen – 20/12/2023
-
Glioblastoma-instructed microglia transit to heterogeneous phenotypic states with phagocytic and dendritic cell-like features in patient tumors and patient-derived orthotopic xenografts – 12/12/2023
-
Bespoke three- photon microscopy and analysis enable deep intravital brain tumor imaging – 10/11/2023
-
Characterizing the GBM cellular landscape by large-scale single-nucleus RNA-sequencing – 10/11/2023
-
Dissecting gbm evolution following standard-of-care by large-scale longitudinal single nucleus RNA-sequencing – 10/11/2023
Related News

Job vacancies
There are no jobs matching this page at the moment. You can view all jobs via the button below.